6.4 Processing data
All data objects that are generated during a sets workflow inherit from the classes from a ‘regular’ workflow. This means that, with some minor exceptions, all of the data processing functionality discussed in the previous chapter (e.g. subsetting, inspection, filtering, plotting, reporting) is also applicable to a sets workflow. For instance, the as.data.table() method can be used for general inspection:
as.data.table(compounds)[1:5, c("group", "score-positive", "score-negative", "compoundName", "set")]#> group score-positive score-negative compoundName set
#> <char> <num> <num> <char> <char>
#> 1: M198_R317_273 3.290700 4.569478 3-(4-chlorophenyl)-1,1-dimethylurea positive,negative
#> 2: M198_R317_273 1.668025 2.025473 3-(3-chlorophenyl)-1,1-dimethylurea positive,negative
#> 3: M198_R317_273 1.594943 1.869558 4-amino-2-chloro-N,N-dimethylbenzamide positive,negative
#> 4: M198_R317_273 1.673759 1.557270 1-(4-chlorophenyl)-3-ethylurea positive,negative
#> 5: M198_R317_273 1.591203 1.865819 3-amino-4-chloro-N,N-dimethylbenzamide positive,negativeIn addition, some the data processing functionality contains additional functionality for a sets workflow:
# only keep feature groups that have positive data
fGroupsPos <- fGroups[, sets = "positive"]
# only keep feature groups that have feature data for all sets
fGroupsF <- filter(fGroups, relMinSets = 1)
# only keep feature groups with features present in both polarities
fGroupsPosNeg <- overlap(fGroups, which = c("positive", "negative"), sets = TRUE)
# only keep feature groups with features that are present only in positive mode
fGroupsOnlyPos <- unique(fGroups, which = "positive", sets = TRUE)And plotting:
plotVenn(fGroups, sets = TRUE) # compare positive/negative features
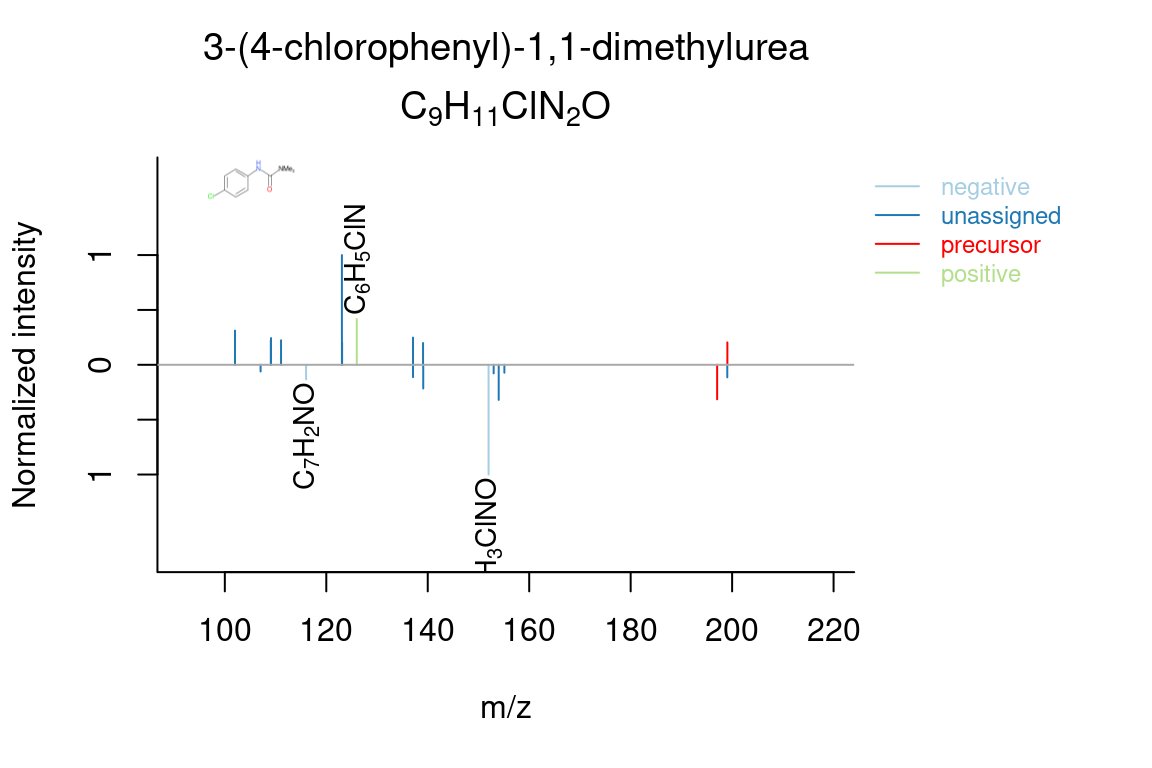
plotSpectrum(compounds, index = 1, groupName = "M198_R317_273", MSPeakLists = mslists,
plotStruct = TRUE)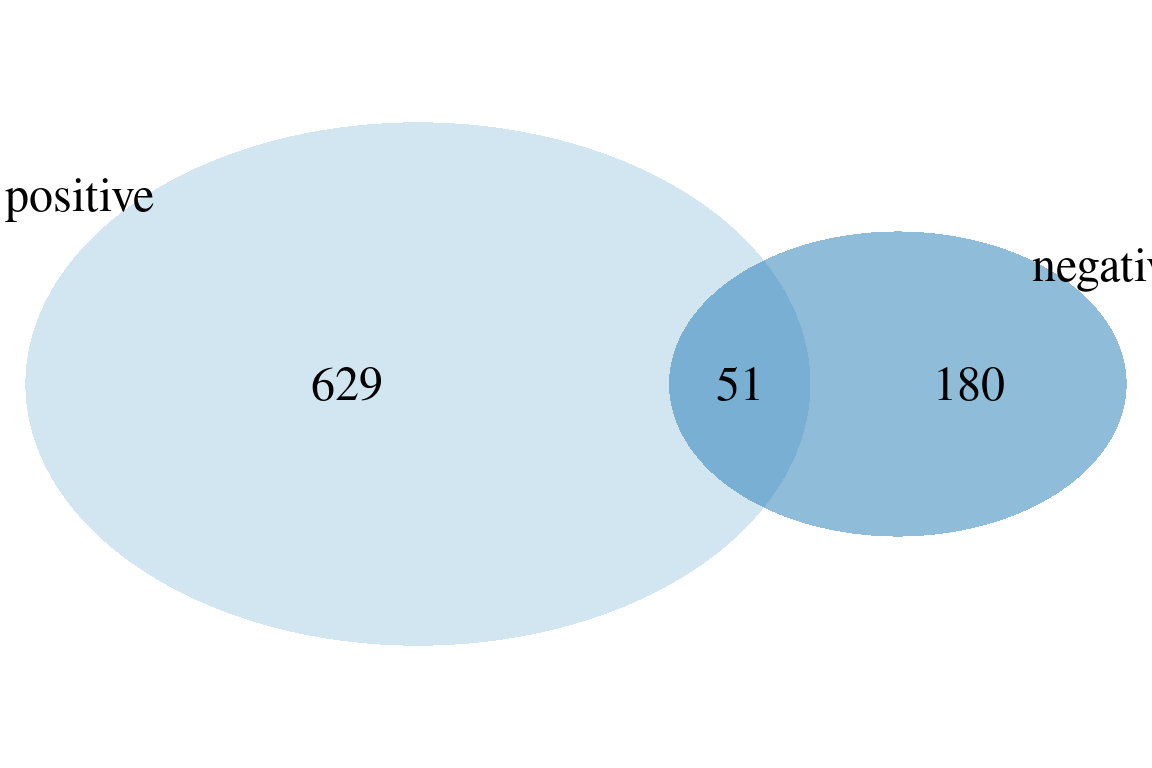
The reference manual for the workflow objects contains specific notes applicable to sets workflows (?featureGroups, ?compounds etc).