7.1 Obtaining transformation product data
The generateTPs function is used to obtain TPs for a particular set of parents. Like other workflow generator functions (findFeatures, generateCompounds), several algorithms are available that do the actual work.
| Algorithm | Usage | Remarks |
|---|---|---|
| BioTransformer | generateTPs(algorithm = "biotransformer", ...) |
Predicts TPs with full structural information |
| CTS | generateTPs(algorithm = "cts", ...) |
Predicts TPs with full structural information |
| Library | generateTPs(algorithm = "library", ...) |
Obtains transformation products from a library (PubChem transformations or custom) |
| Formula library | generateTPs(algorithm = "library_formula", ...) |
Obtains transformation products from a library (only formula data) |
| Metabolic logic | generateTPs(algorithm = "logic", ...) |
Uses pre-defined logic to predict TPs based on common elemental differences (e.g. hydroxylation, demethylation). Based on Schollee et al. (2015). |
The output of these algorithms can be distinguished in three categories:
- Structural TPs (
biotransformer,ctsandlibrary) come with full structural information for the TPs (e.g. formula, SMILES, predicted Log P). As such, the corresponding algorithms also require the full chemical structure of the parent compound. - Formula TPs (
library_formula) are similar to structural TPs, but only involve formula and no other structural information. - Calculated TPs (
logic) are based solely on m/z differences and only require the feature masses.
Algorithms that fall into the first category are typically used when parents are known in advance, for instance, from a target or suspect screening. This is also true for the second category, however, here only formula data is used, which is useful when the complete structure of parents and/or TPs are not known. Calculated TPs allow TP prediction for all features, even when nothing is known about their structure. This is most suitable for full non-target analysis, however, extra care must be taken to rule out false positives. Finally, the logic used to calculate TPs can also be used to automatically to generate a library suitable for the library_formula algorithm, which allows a hybrid approach of the second and third categories.
An overview of common arguments for TP generation is listed below.
| Argument | Algorithm(s) | Remarks |
|---|---|---|
parents |
biotransformer, cts, library |
The input parents. See section below. |
fGroups |
logic |
The input feature groups to calculate TPs for. |
type |
biotransformer |
The prediction type: "env", "ecbased", "cyp450", "phaseII", "hgut", "superbio", "allHuman". See BioTransformer for more details. |
transLibrary |
cts |
The transformation library that should be used: "hydrolysis", "abiotic_reduction", "photolysis_unranked", "photolysis_ranked", "mammalian_metabolism", "combined_abioticreduction_hydrolysis", "combined_photolysis_abiotic_hydrolysis". See cts for more details. |
TPLibrary/transformations |
library/logic |
Custom TP library/transformation rules. |
generation |
biotransformer, cts, library |
The amount of transformation generations to predict. |
adduct |
logic |
The assumed adduct of the parents (e.g. "[M+H]+"). Not needed when adduct annotations are available. |
calcSims |
biotransformer, cts, library |
If TRUE then structural similarities between the parent and TPs is calculated, which can be useful for post-processing (discussed later). |
7.1.1 Parent input
The input parent structures to generate structural/formula TPs (biotransformer, cts, library and library_formula algorithms) must be specified as one of the following:
- A suspect list (follows the same format as suspect screening)
- A feature groups object with screening results (e.g. obtained with
screenSuspects, see suspect screening) - A
compoundsobject obtained with compound annotation (not supported forlibrary_formula)
In the former two cases the parent information is taken from the suspect list or from the hits in a suspect screening worklow, respectively. The last case is more suitable for when the parents are not completely known. In this case, the candidate structures from a compound annotation are used as input to obtain TPs. Since all the candidates are used, it is highly recommend to filter the object in advance, for instance, with the topMost filter. For library and library_formula, the parent input is optional: if no parents are specified then TP data for all parents in the database is used.
For the logic algorithm TPs are predicted directly for feature groups. Since this algorithm can only perform very basic validity checks, it is strongly recommended to first prioritize the feature group data.
Some typical examples:
# predict environmental TPs with BioTransformer for all parents in a suspect list
TPsBT <- generateTPs("biotransformer", parents = patRoonData::suspectsPos,
type = "env")
# obtain all TPs from the default library
TPsLib <- generateTPs("library")
# get TPs for the parents from a suspect screening
TPsLib <- generateTPs("library", parents = fGroupsScr)
# calculate TPs for all feature groups
TPsLogic <- generateTPs("logic", fGroups, adduct = "[M+H]+")7.1.2 Processing data
Similar to other workflow data, several generic functions are available to inspect the data:
| Generic | Remarks |
|---|---|
length() |
Returns the total number of transformation products |
names() |
Returns the names of the parents |
parents() |
Returns a table with information about the parents |
products() |
Returns a list with for each parent a table with TPs |
as.data.table(), as.data.frame |
Convert all the object information into a data.table/data.frame |
"[[" / "$" operators |
Extract TP information for a specified parent |
Some examples:
# just show a few columns in this example, there are many more!
# note: the double dot syntax (..cols) is necessary since the data is stored as data.tables
cols <- c("name", "formula", "InChIKey")
parents(TPs)[1:5, ..cols]#> name formula InChIKey
#> <char> <char> <char>
#> 1: DEET C12H17NO MMOXZBCLCQITDF-UHFFFAOYSA-N
#> 2: Irgarol C11H19N5S HDHLIWCXDDZUFH-UHFFFAOYSA-N
#> 3: Prometryne C10H19N5S AAEVYOVXGOFMJO-UHFFFAOYSA-N
#> 4: Trimethoprim C14H18N4O3 IEDVJHCEMCRBQM-UHFFFAOYSA-N
#> 5: 1H-benzotriazole C6H5N3 QRUDEWIWKLJBPS-UHFFFAOYSA-N#> name formula InChIKey
#> <char> <char> <char>
#> 1: DEET-TP1 C12H17NO2 FRZJZRVZZNTMAW-UHFFFAOYSA-N
#> 2: DEET-TP2 C12H17NO2 KVTUZBGZTRABBQ-UHFFFAOYSA-N
#> 3: DEET-TP3 C2H4O IKHGUXGNUITLKF-UHFFFAOYSA-N
#> 4: DEET-TP4 C10H13NO FPINATACRXASTP-UHFFFAOYSA-N
#> 5: DEET-TP4 C10H13NO FPINATACRXASTP-UHFFFAOYSA-N
#> 6: DEET-TP5 C4H11N HPNMFZURTQLUMO-UHFFFAOYSA-N
#> 7: DEET-TP6 C8H7O2 GPSDUZXPYCFOSQ-UHFFFAOYSA-M
#> 8: DEET-TP7 C8H9NO WGRPQCFFBRDZFV-UHFFFAOYSA-N
#> 9: DEET-TP1 C12H17NO2 FRZJZRVZZNTMAW-UHFFFAOYSA-N#> name formula InChIKey
#> <char> <char> <char>
#> 1: Irgarol-TP1 C8H15N5S MWWBDLRPMWTLRX-UHFFFAOYSA-N
#> 2: Irgarol-TP2 C11H19N5OS HFCMSBLJLJOGGL-UHFFFAOYSA-N#> parent transformation name_lib
#> <char> <char> <char>
#> 1: DEET Aliphatic hydroxylation of methyl carbon adjacent to aromatic ring / Human Phase I N,N-diethyl-m-hydroxymethylbenzamide
#> 2: DEET Hydroxylation of terminal methyl / Human Phase I N-Ethyl-N-(2-hydroxyethyl)-3-methylbenzamide
#> 3: DEET N-dealkylation of tertiary carboxamide / Human Phase I acetaldehyde
#> 4: DEET N-dealkylation of tertiary carboxamide / Human Phase I N-ethyl-m-toluamide
#> 5: DEET Metabolism Benzamide, N-ethyl-3-methyl-In addition, the following generic functions are available to modify or convert the object data:
| Generic | Classes | Remarks |
|---|---|---|
"[" operator |
All | Subset this object on given parents |
filter |
All | Filters this object |
convertToSuspects |
All | Generates a suspect list of all TPs (and optionally parents) that is suitable for screenSuspects |
TPs2 <- TPs[1:10] # only keep results for first ten parents
# only keep TPs with likely/probably likelihood (specific property for CTS algorithm)
TPsF <- filter(TPs, properties = list(likelihood = c("LIKELY", "PROBABLE")))
# do a suspect screening for all TPs and their parents
suspects <- convertToSuspects(TPs, includeParents = TRUE)
fGroupsScr <- screenSuspects(fGroups, suspects, onlyHits = TRUE)The convertToSuspects function is always part of a workflow, and is discussed further in the next section.
7.1.2.1 Structural TPs specifics
For structural TPs several additional generic functions are available:
| Generic | Remarks |
|---|---|
filter |
Filters this object (additional functionality for structural TPs) |
convertToMFDB |
Generates a MetFrag database for all TPs (and optionally parents) |
plotGraph |
Generates an interactive plot to explore transformation hierarchies |
plotVenn, plotUpSet |
Compare results between different algorithms with Venn/UpSet diagrams |
The convertToMFDB function is especially handy with predicted TPs, as it allows generating a compound database for TPs that may not be available in commonly used databases. This is further demonstrated in the first example.
# remove transformation products that are isomers to their parent or sibling TPs
# may simplify data as these are often difficult to identify
TPsF <- filter(TPs, removeParentIsomers = TRUE, removeTPIsomers = TRUE)
# remove duplicate transformation products from each parent
# these can occur if different pathways yield the same TPs
TPsF <- filter(TPs, removeDuplicates = TRUE)
# only keep TPs that have a structural similarity to their parent of >= 0.5
# (needs calcSims=TRUE when executing generateTPs())
TPsF <- filter(TPs, minSimilarity = 0.5)
# use the TP data for a specialized MetFrag database
convertToMFDB(TPs, "TP-database.csv", includeParents = FALSE)
compoundsTPs <- generateCompounds(fGroups, mslists, "metfrag", database = "csv",
extraOpts = list(LocalDatabasePath = "TP-database.csv"))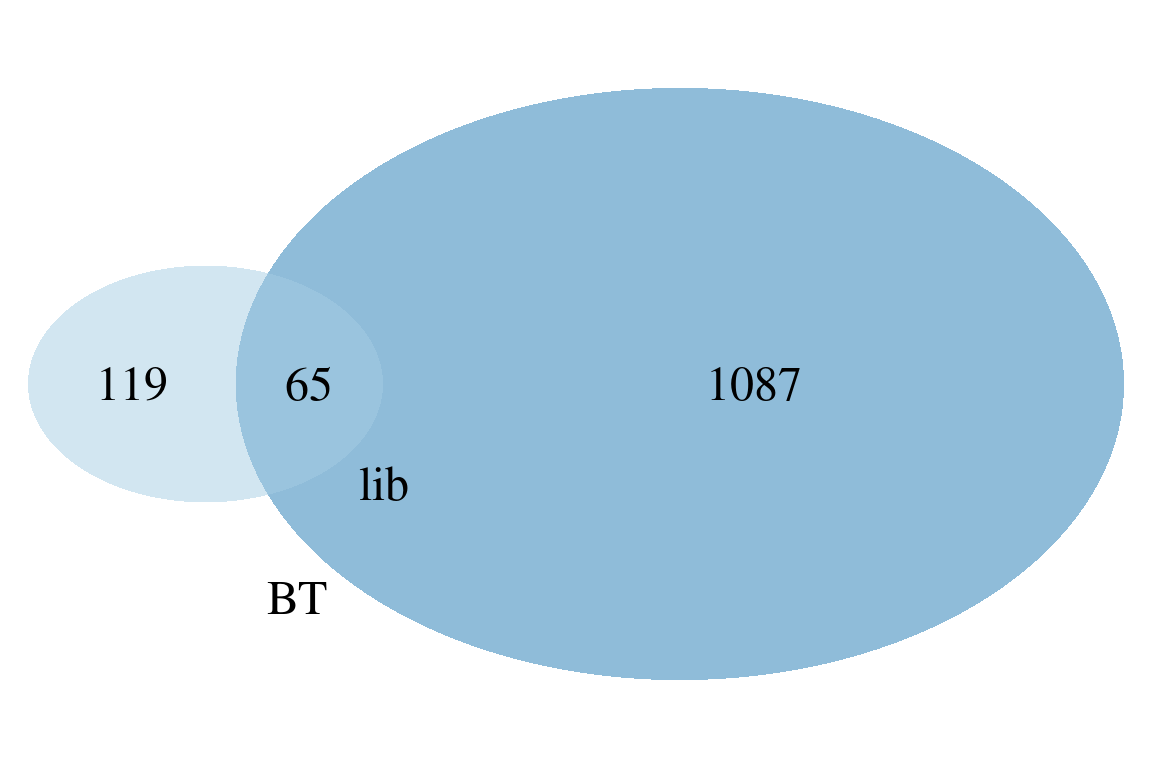
Finally, results from different algorithms can be combined with the consensus generic function. This is further discussed in algorithm consensus.
7.1.3 (Custom) Libraries and transformations
By default the library and logic algorithms use data that is installed with patRoon (based on PubChem transformations and Schollee et al. (2015), respectively). However, it is also possible to use custom data. For the library_formula no default library is provided, however, these can easily be generated as is discussed at the end of the section.
To use a custom TP structure library a simple data.frame is needed with the names, SMILES and optionally log P values for the parents and TPs. The log P values are used for prediction of the retention time direction of a TP compared to its parent, as is discussed further in the next section. The following small library has two TPs for benzotriazole and one for DEET:
myTPLib <- data.frame(parent_name = c("1H-Benzotriazole", "1H-Benzotriazole", "DEET"),
parent_SMILES = c("C1=CC2=NNN=C2C=C1", "C1=CC2=NNN=C2C=C1", "CCN(CC)C(=O)C1=CC=CC(=C1)C"),
TP_name = c("1-Methylbenzotriazole", "1-Hydroxybenzotriazole", "N-ethyl-m-toluamide"),
TP_SMILES = c("CN1C2=CC=CC=C2N=N1", "C1=CC=C2C(=C1)N=NN2O", "CCNC(=O)C1=CC=CC(=C1)C"))
myTPLib#> parent_name parent_SMILES TP_name TP_SMILES
#> 1 1H-Benzotriazole C1=CC2=NNN=C2C=C1 1-Methylbenzotriazole CN1C2=CC=CC=C2N=N1
#> 2 1H-Benzotriazole C1=CC2=NNN=C2C=C1 1-Hydroxybenzotriazole C1=CC=C2C(=C1)N=NN2O
#> 3 DEET CCN(CC)C(=O)C1=CC=CC(=C1)C N-ethyl-m-toluamide CCNC(=O)C1=CC=CC(=C1)CTo use this library, simply pass it to the TPLibrary argument:
For library_formula the library follows the same format. However, here the formula should be specified instead of the SMILES with the parent_formula and TP_formula columns (although it is still allowed to only specify SMILES, as in this case the formulae are automatically calculated).
For the logic algorithm a table with custom transformation rules can be specified for TP calculations:
myTrans <- data.frame(transformation = c("hydroxylation", "demethylation"),
add = c("O", ""),
sub = c("", "CH2"),
retDir = c(-1, -1))
myTrans#> transformation add sub retDir
#> 1 hydroxylation O -1
#> 2 demethylation CH2 -1The add and sub columns are used to denote the elements that are added or subtracted by the reaction. These are used to calculate mass differences between parents and TPs. The retDir column is used to indicate the retention time direction of the parent compared to the TP: -1 (elutes before parent), 1 (elutes after parent) or 0 (similar or unknown). The next section describes how this data can be used to filter TPs. The custom rules can be used by passing them to the transformations argument:
The genFormulaTPLibrary() utility function can be used to automatically generate TP libraries suitable for the library_formula algorithm. The transformation rules to calculate TPs are specified in the same format as used by the logic algorithm.
myTPFormLib <- genFormulaTPLibrary(parents = patRoonData::suspectsPos, transformations = myTrans)
# also calculate second generation TPs (TPs of TPs)
myTPFormLib2 <- genFormulaTPLibrary(parents = patRoonData::suspectsPos, transformations = myTrans,
generations = 2)
# Use library
TPs <- generateTPs("library_formula", TPLibrary = myTPFormLib)Compared to the logic algorithm, the library_formula algorithm is more (and only) suitable for suspect/target screening workflows, allows multiple transformation generations and allows better customization through manually adding/removing TPs from the library prior to passing it to generateTPs().